Cross-Modality Domain Adaptation for Medical Image Segmentation and Classification
Unsupervised 3D Semantic Segmentation and Classification Domain Adaptation
👋 2022: Results are out! The crossMoDA 2021 paper is now on ArXiv

Announcements
Aim
Domain Adaptation (DA) has recently raised strong interests in the medical imaging community. By encouraging algorithms to be robust to unseen situations or different input data domains, Domain Adaptation improves the applicability of machine learning approaches to various clinical settings. While a large variety of DA techniques has been proposed, most of these techniques have been validated either on private datasets or on small publicly available datasets. Moreover, these datasets mostly address single-class problems. To tackle these limitations, the crossMoDA challenge introduced the first large and multi-class dataset for unsupervised cross-modality Domain Adaptation. Compared to the previous crossMoDA instance, which made use of single-institution data and featured a single segmentation task, the 2022 edition extends the segmentation task by including multi-institutional data and introduces a new classification task.
Tasks
Participants are free to choose whether they want to focus only on one or both tasks.
Task 1: Vestibular schwannoma and cochlea segmentation
The goal of the segmentation task (Task 1) is to segment two key brain structures (tumour and cochlea) involved in the follow-up and treatment planning of vestibular schwannoma (VS). The segmentation of these two structures is required for radiosurgery, a standard VS treatment. Moreover, tumour volume measurement has also been shown to be the most accurate measurement for evaluating VS growth. The diagnosis and surveillance in patients with VS are commonly performed using contrast-enhanced T1 (ceT1) MR imaging. However, there is growing interest in using non-contrast imaging sequences such as high-resolution T2 (hrT2) imaging, as it mitigates the risks associated with gadolinium-containing contrast agents. Furthermore, in addition to improving patient safety, hrT2 imaging is 10 times more cost-efficient than ceT1 imaging.
For this reason, we proposed an unsupervised cross-modality segmentation benchmark (from ceT1 to hrT2) that aims to perform VS and cochlea segmentation on hrT2 scans automatically. The training source and target sets are respectively unpaired annotated ceT1 and non-annotated hrT2 scans from both pre-operative and post-operative time points. To validate the robustness of the proposed approaches on different hrT2 settings, multi-institutional scans from centres in London, UK and Tilburg, NL are used in this task.
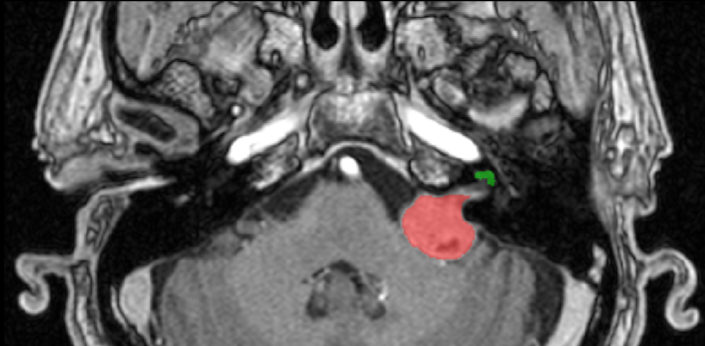 Source (contrast-enhanced T1)
Source (contrast-enhanced T1)
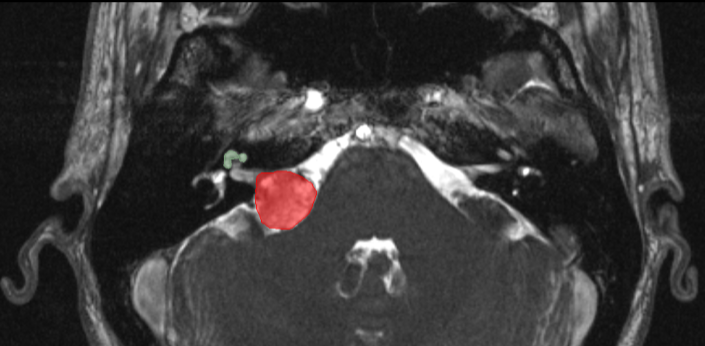 Target (high resolution T2)
Target (high resolution T2)
Task 2: Koos classification
The goal of the classification task (Task 2) is to automatically classify hrT2 images with VS according to the Koos grade. The Koos grading scale is a classification system for VS that characterises the tumour and its impact on adjacent brain structures (e.g., brain stem, cerebellum). The Koos classification is commonly determined to decide on the treatment plan (surveillance, radiosurgery, open surgery). Similarly to the VS segmentation, Koos grading is currently performed on ceT1 scans, but hrT2 could be used. For this reason, we propose an unsupervised cross- modality classification benchmark (from ceT1 to hrT2) that aims to determine the Koos grade on hrT2 scans automatically. Only pre-operative data is used for this task. Again, multi-institutional scans from centres in London, UK and Tilburg, NL are used in this task.
Data
Participants are free to use the data from one task for the other task.
London data
All images were obtained on a 32-channel Siemens Avanto 1.5T scanner using a Siemens single-channel head coil:
- Contrast-enhanced T1-weighted imaging was performed with an MPRAGE sequence with in-plane resolution of 0.4×0.4mm, in-plane matrix of 512×512, and slice thickness of 1.0 to 1.5 mm (TR=1900 ms, TE=2.97 ms, TI=1100 ms)
- High-resolution T2-weighted imaging was performed with a 3D CISS or FIESTA sequence in-plane resolution of 0.5x0.5mm, in-plane matrix of 384x384 or 448x448, and slice thickness of 1.0 to 1.5 mm (TR=9.4 ms, TE=4.23ms).
Tilburg data
All images were obtained on a Philips Ingenia 1.5T scanner using a Philips quadrature head coil:
- Contrast enhanced T1-weighted imaging was performed with a 3D-FFE sequence with in-plane resolution of 0.8×0.8mm, in-plane matrix of 256×256, and slice thickness of 1.5 mm (TR=25 ms, TE=1.82 ms).
- High-resolution T2-weighted imaging was performed with a 3D-TSE sequence with in-plane resolution of 0.4x0.4mm, in-plane matrix of 512×512, and slice thickness of 1.0 mm (TR=2700 ms, TE=160 ms, ETL=50).
All data will be made available online with a permissive non-commercial copyright-license (CC BY-NC-SA 4.0), allowing for data to be shared, distributed and improved upon. All structures were manually segmented in consensus by the treating neurosurgeon and physicist using both the ceT1 and hrT2 images. To cite this data, please refer to https://arxiv.org/abs/2201.02831.
Rules
No additional data is allowed, including the data released on TCIA and pre-trained models. The use of a generic brain atlas is tolerated as long as its use is made clear and justified.
Example of tolerated use cases:- Spatial normalisation to MNI space
- Use of classical single-atlas based tools (e.g., SPM)
Examples of cases that are not allowed:
- Multi-atlas registration based approaches in the target domain
No additional annotations are allowed.
Models can be adapted (trained) on the target domain (using the provided target training set) in an unsupervised way, i.e. without labels.
The participant teams will be required to release their training and testing code and explain how they fine-tuned their hyper-parameters. Note that the code can be shared with the organisers only as a way to verify validity, and if needed, NDAs can be signed.
The top 3 ranked teams will be required to submit their training and testing codes in a docker container for verification after the challenge submission deadline in order to ensure that the challenge rules have been respected.
Evaluation
Task 1: Vestibular schwannoma and cochlea segmentation
Classical semantic segmentation metrics, in this case, the Dice Score (DSC) and the Average Symmetric Surface Distance (ASSD), will be used to assess different aspects of the performance of the region of interest. These metrics are implemented here. The metrics (DSC, ASSD) were chosen because of their simplicity, their popularity, their rank stability, and their ability to assess the accuracy of the predictions.
Participating teams are ranked for each target testing subject, for each evaluated region (i.e., VS and cochlea), and for each measure (i.e., DSC and ASSD). The final ranking score for each team is then calculated by firstly averaging across all these individual rankings for each patient (i.e., Cumulative Rank), and then averaging these cumulative ranks across all patients for each participating team.
Task 2: Koos classification
Participating teams are ranked based on their Macro-averaged mean absolute error (MA-MAE). Macro-averaged mean absolute error is well-designed for ordinal and imbalanced classification problems. It has been used successfully in other challenges such as SemEval-2017.
Timeline
| 2nd May 2022: | Release of the training and validation data. |
| 12th May 2022: | Start of the validation period. |
| 5th August 2022: | Start of the evaluation period. |
| 15th August 2022: | End of the evaluation period. |
| 18th September 2022: | Challenge results are announced at MICCAI 2022. |
| 30th November 2022: | Participants are invited to submit their methods to the MICCAI 2022 BrainLes Workshop. |
| December 2022: | Submission of a joint manuscript summarizing the results of the challenge to a high-impact journal in the field. |
Winners of crossMoDA-2022 challenge
Methods of the top-performing 2022 teams are described here.
Task 1: Segmentation
| # | Team Name | Affiliation | DSC | Technical report |
|---|---|---|---|---|
| 1 | ne2e | Peking University, Beijing, China | 86.9 | Unsupervised Domain Adaptation in Semantic Segmentation Based on Pixel Alignment and Self-Training (PAST) |
| 2 | MAI | Korea University, Korea | 86.8 | Multi-view Cross-Modality MR Image Translation for Vestibular Schwannoma and Cochlea Segmentation |
| 3 | LaTIM | Inserm, France | 85.9 | Tumor blending augmentation using one-shot generative learning for vestibular schwannoma and cochlea cross-modal segmentation |
Task 2: Koos Classification
| # | Team Name | Affiliation | MA-MAE | Technical report |
|---|---|---|---|---|
| 1 | SJTU_EIEE_2 | Shanghai Jiao Tong University, China | 0.26 | Koos Classification of Vestibular Schwannoma via Image Translation-Based Unsupervised Cross-Modality Domain Adaptation |
| 2 | Super Polymerization | Radboud University, the Netherlands | 0.37 | Unsupervised Cross-Modality Domain Adaptation for Vestibular Schwannoma Segmentation and Koos Grade Prediction based on Semi-Supervised Contrastive Learning |
| 3 | skjp | Muroran Institute of Technology, Japan | 0.84 | Unsupervised Domain Adaptation for MRI Volume Segmentation and Classification Using Image-to-Image Translation |